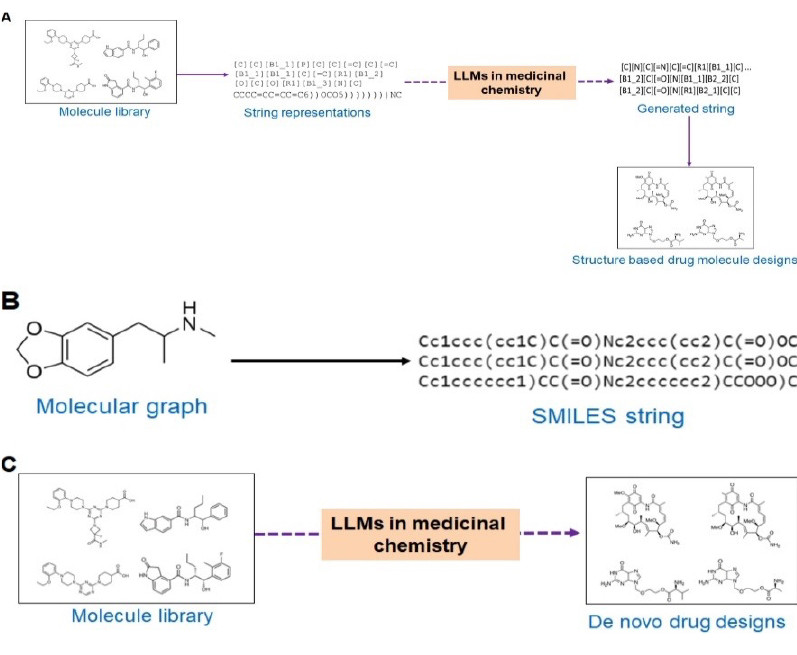
Source: [https://doi.org/10.1016/j.jare.2025.02.011]. Overview of the application of LLMs for drug design. (a) the application of LLMs in structure-based drug molecule design, (b) the generation of SMILES strings from molecular graphs during drug design, and (c) the application of LLMs in de novo drug design.
DrugLLM uses open LLM and GMR to produce accurate molecular representation using a few-shot processes. DrugLLM successfully generated bioactive HCN2 inhibitors, which were verified in laboratory settings. MolGPT is another generative model designed for molecular generation using a transformer-decoder architecture. Ye et al. developed an LLM model, DrugAssist, to optimise the drug molecule by grasping the underlying patterns in chemical structures. Recently, another research group developed an LLM for drug design and understanding three-dimensional (3D) structures via tokenisation.
3. De novo drug design
Researchers are also developing drug molecules through de novo drug design. A conditional GPT-based model cMolGPT, employs multihead attention mechanisms to conditionally generate SMILES strings by identifying closely matched molecules of real target-specific compounds. Another multi-objective LLM model, FSM-DDTR, performs de novo drug design using transformer-based architecture to explore the vast chemical space and generate drug molecules with all optimal drug-likeness properties.
Another useful tool is DrugChat, an AI-powered conversational agent. It can handle various types of data inputs and generate comprehensive outputs, which helps in understanding the structure-activity relationships and is useful for lead optimisation, drug repurposing, etc.
4. Biomarker identification
Identifying biomarkers is a critical step in drug discovery. According to a review of 1,079 oncology drugs, 24% of biomarker-based treatments showed success rate, compared to 6% for non-biomarker-based compounds. LLMs excel in this area due to their ability to process and analyse vast datasets. By rapidly reviewing thousands of research papers, clinical reports and genomic databases, LLMs uncover hidden patterns and associations, accelerating biomarker discovery.
BRAD is another example of an agentic system that combines LLMs with external tools and data to streamline research workflows. This workflow utilises user-provided gene expression (RNA-seq) data along with research literature as input, conducts both biomarker selection and enrichment analysis, and generates a report that interprets the data. There are a few cases where this agentic system is used to discover biomarkers that indicate disease progression and response to the treatment of neurodegenerative diseases. Some researchers already used LLMs to identify gene signatures associated with diseases like ulcerative colitis.
5. Multi-LLMs-based intelligent agent frameworks in drug development
The use of multi-LLMs-based intelligent agents in drug discovery is a cutting-edge approach that leverages the power of large language models to automate and enhance various stages of the drug discovery process. One of the most important concepts in drug discovery is “drug repurposing,” which helps us to find a molecule for a symptom that already exists as a drug to treat other diseases and conditions. A popular drug repurposing LLM model is DrugReAlign, which uses multisource prompt techniques developed by a collaborative team of researchers. Researchers from USC, CMU and RPI developed DrugAgent, a multi-agent framework that marks a significant advancement in leveraging LLMs for automating critical aspects of drug discovery. It automates tasks such as data acquisition, model selection, drug-disease interactions and evaluation.
Another popular multi-LLM-based intelligent agent is Coscientist, developed by Google's AI unit DeepMind, capable of automating the design, planning, writing and executing code, and manipulating robotic experimental platforms and performing complex scientific experiments, such as compound synthesis. Coscientist has demonstrated its capabilities by successfully synthesising complex compounds like ibuprofen and nitroaniline.
Real-world applications of LLMs in the industry in drug discovery
These real-world applications of LLMs highlight the potential of AI in drug discovery and development, providing a glimpse into a future shaped by these advanced technologies. Few real-world applications in the industry are listed below:
- BioNTech and COVID-19 vaccine development: BioNTech utilised LLMs to develop the first mRNA-based COVID-19 vaccine with 95% efficacy, at multiple stages of its COVID-19 vaccine development
- MegaMolBART by NVIDIA: MegaMolBART employs an LLM model developed for small molecule drug discovery and cheminformatics, utilising the SMILES notation and the NeMo-Megatron framework
- Meta’s ESM-2 LLM: This application can help design atomic-level new proteins, without the need for multiple sequence alignments
- Pfizer's use of LLMs: Pfizer's in-house LLM, Charlie, is used to analyse vast amounts of data from various sources, including clinical trial results, patient records and scientific literature
Cognizant’s capabilities towards LLM-based applications on drug discovery
Cognizant has already developed a gen AI-based drug discovery platform and other bioinformatics/cheminformatics software using cloud technology for a leading pharmaceutical company in Germany. Recently, Cognizant has engaged in developing an autonomous virtual laboratory LLM-based platform for the multinational pharmaceutical industry. This platform will have several applications, including creating small molecules, optimising chemical reactions, predicting solubility and predicting reaction mechanisms. The Cognizant teams are analysing the data and proposing new ideas to improve the efficiency of the platform. Our teams are collaborating with NVIDIA to advance drug discovery by using generative AI models that help in designing novel small molecules, virtual screening, predict complex protein structures and even forecast the subcellular localisation of protein sequences. Our dedicated research and development team is actively leading transformation by improving wet lab experiments with machine learning models, enabling lab automation and upgrading laboratory information management systems (LIMS). We have also employed a language model for TCRs to predict TCR-epitope binding across various human leukocyte antigen (HLA) class-I types.
Conclusions
Large language models are transforming the pharmaceutical industry by significantly enhancing the drug discovery process. These advanced AI tools can rapidly analyse vast amounts of scientific data, identify patterns and propose novel drug candidates with high precision. Overall, the integration of LLMs into pharmaceutical research is revolutionising the industry, making drug discovery faster, more accurate and cost-effective.